
Each item in the search menu, and each item in the context (right-click) menu, is backed by one SPARQL query. In the background, the General SPARQL app maintains a list of SPARQL queries. Watch this screencast and it will start to make sense: And you can continue this process, jumping from one entity to the next.
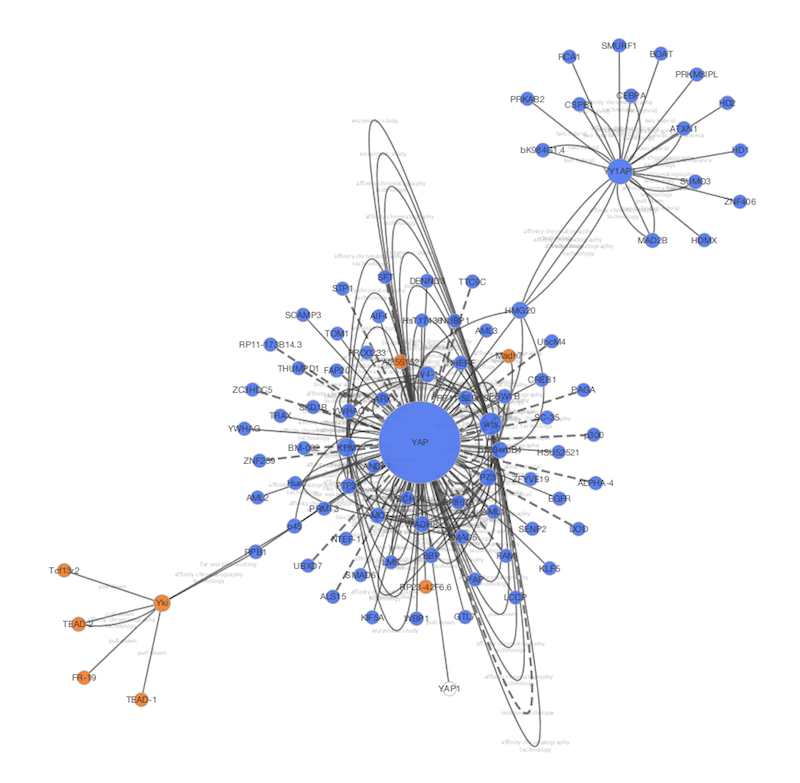
Or the gene that encodes for your protein. Or all the reactions that your protein is part of. For example, all the pathways that are related to your protein. You can then right-click on this node to pull in related entities. The app places a single node in your network. Edges can represent any type of relation between those entities.įor example, you can start by searching for a protein of interest. Nodes can represent various biological entities, such as: a pathway, a protein, a reaction, or a compound. Nodes and edges can be added to a network in a piecemeal fashion. The app lets you build a network step by step. The General SPARQL app is one of the new ways to present triple data. We shouldn’t expect scientists to write SPARQL queries anymore than we expect them to carry adjustable pliers to a restroom visit. The problem is that SPARQL is supposed to be only a piece of plumbing at the bottom of a software stack. The problem isn’t that SPARQL is hard to use per se (it’s really rather plain and sensible).
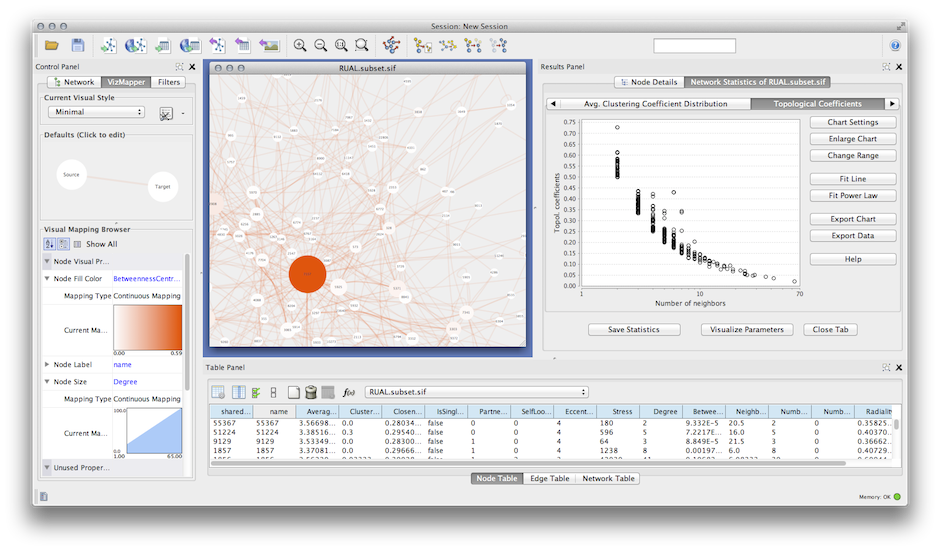
Integrating data is only half of the problem, we also have to present that data.
#Cytoscape 3 increase memory how to#
Triple stores are great for data integration, but you still have to figure out how to put that data in the hands of scientists. But how do we put that data in the hands of scientists?Īt General Bioinformatics we put data in triple stores, and use SPARQL to query that data. We can now easily solve the problem of bioinformatics data integration.


 0 kommentar(er)
0 kommentar(er)
